A pictorial representation of the PIZSA algorithm.
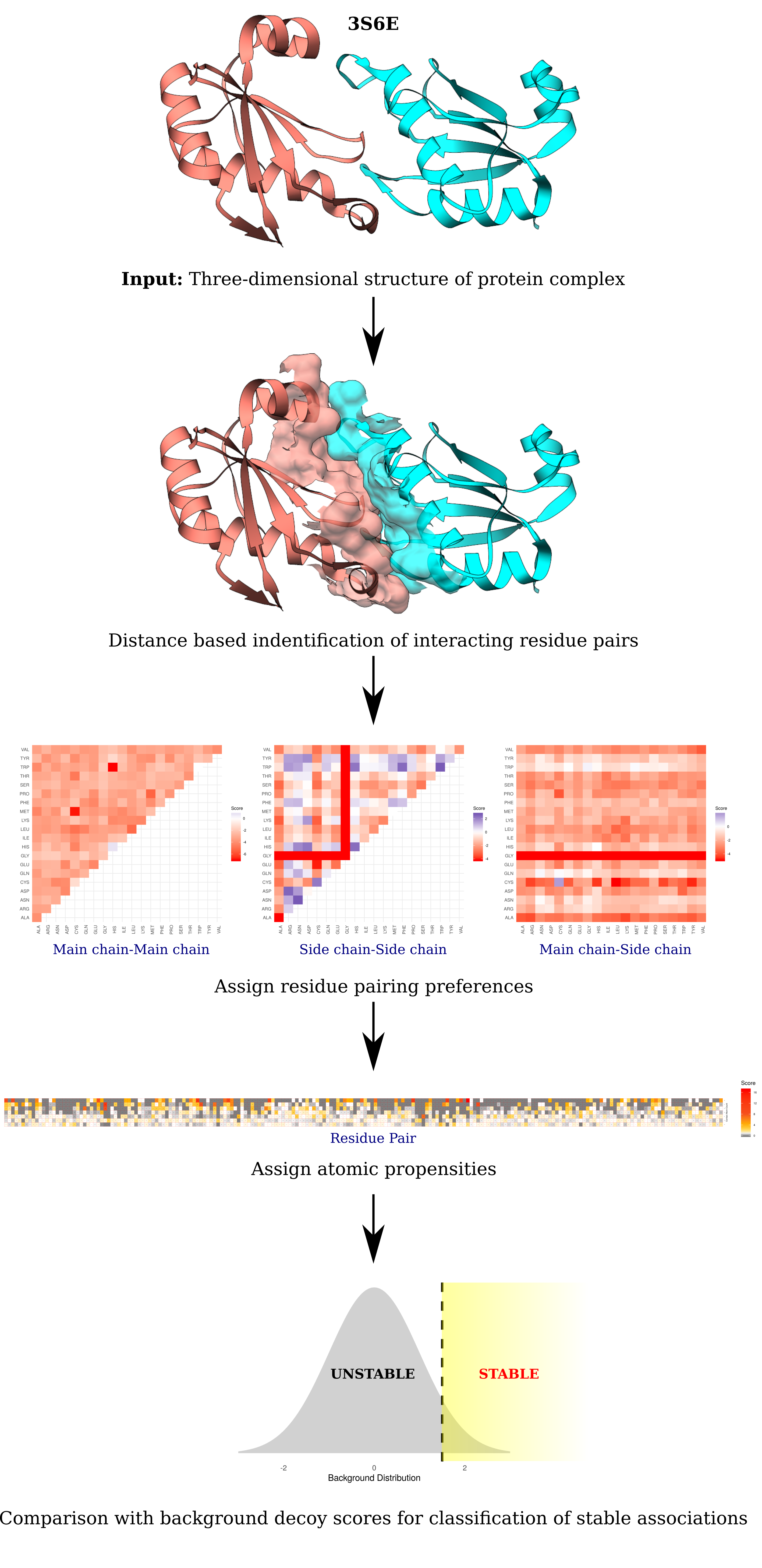
Interacting residue pairs on protein complexes are identified based on a distance threshold. Residues from different chains that have at least a single heavy atom each within a distance threshold of each other are identified as interacting residue pairs. Every residue pair acquires three different scores based on its type of interaction (main chain - main chain, main chain - side chain and side chain - side chain). These scores are further normalized with atomic propensities that quantify the likelihood of the interaction being mediated by a specific number of atoms. The interaction receives a score that is the sum of all residue pair scores. Protein complexes are assigned a Z Score by comparing their scores to the pre-calculated background scores of a set of non-native interactions. The protein complex is classified as a stable association if it receives a score greater than the Z Score threshold.