

Temperature-sensitive (Ts) mutants of a gene are ones in which there is a marked drop in the level or activity of the gene product when the gene is expressed above a certain temperature (non-permissive temperature). Below this temperature, the activity of the mutant is similar to that of the wild type.
Ts mutants are powerful tools to study protein function in vivo and in cell culture. They provide a reversible mechanism to lower the level of a specific gene product at any stage in the growth of the organism simply by changing the temperature of growth. Ts mutants are widely used to reversibly modulate protein function in vivo and to understand functions of essential genes. Despite this, little is known about the protein structural features and mechanisms responsible for generating a Ts phenotype. Also, such mutants are often difficult to isolate by conventional experimental techniques, limiting their use.
|
|
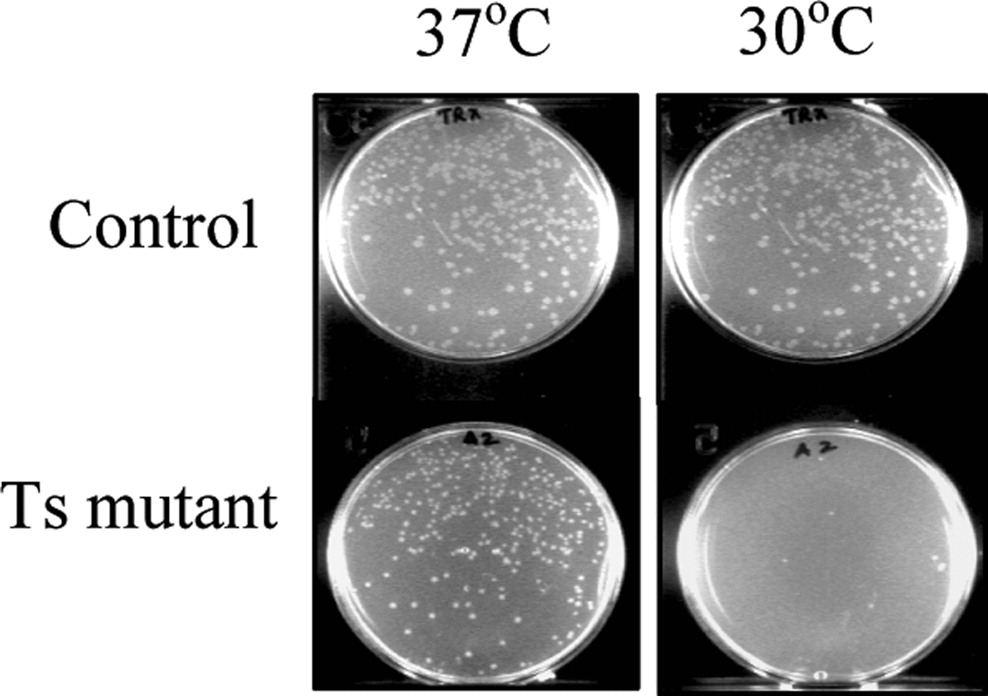
The protein Controller for cell death-B (CcdB) in e. coli when expressed results in the death of the bacterium. The designed Ts mutants of CcdB are only functional at lower temperatures. (Reference/Image Source: Bajaj et al, 2008) |
|
|
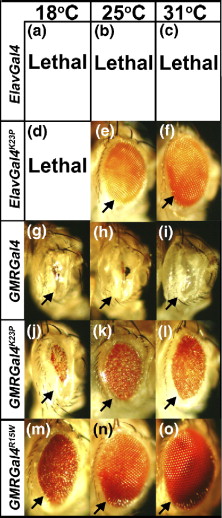
Drosophila eye phenotypes observed when apoptotic UAS-hid reporter is expressed under control of designed temperature-sensitive GMR and Elav wt/mutant Gal4 drivers at different temperature.The arrow points to a similar position in all the eye photographs. (Reference/Image Source: Mondal et al, 2007) |
The server attempts to identify a small set of amino acid residues in a query protein that have a high probability of being buried (side-chain accessibilities less than 5%, expressed in terms of residue depth). The server suggests substitutions at these buried positions that are most likely to result in a Ts phenotype.